Calculating multidimensional Site Frequency Spectra for Demographic Analyses
Many demographic analyses require Site Frequency Spectra (SFS) for their workflow. Different formats and types complicate a straight-forward approach and compatibilty. In this blog post, I introduce you to SFS, their inference, conversion, and use-cases.
BIOINFORMATICS
Tim L. Heller
2/16/20253 min read
Introduction
Site Frequency Spectra (SFS), also known as Allele Frequency Spectra, are a simplified representation of genomic data that enable efficient and accurate inference of various demographic statistics. They show the distribution of allele frequencies across biallelic loci in a population, typically focusing on the frequencies of derived alleles.
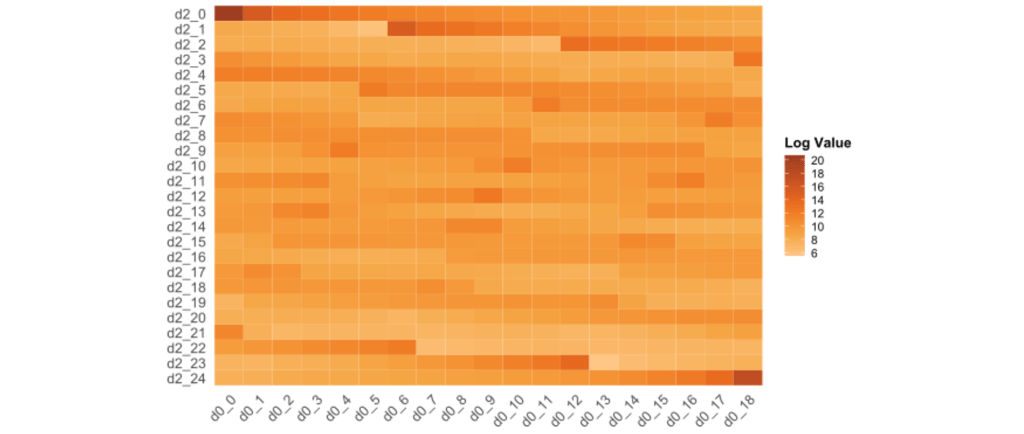
Numerous algorithms and tools in population genomics fully depend on this type of data, as other formats may require too many ressources. In population genomics, we are predominantly interested in the difference between two or more populations, thus, we often need to call multidimensional SFS, such as 2dSFS or even higher. Calling 1dSFS is usually just required as an intermediate step to multidimensional SFS. Two-dimensional SFS can be understood as a two-dimensional array, as depicted in the heatmap accompanying this tutorial. Three-dimensional SFS expand this interpretation by the third dimension, adding slices of 2dSFS for each of the member of the third population.
Types of SFS
There are two distinct types of SFS.
The derived (unfolded) SFS counts the derived mutations relative to an ancestral state across the samples. This method is favoured if a closely related outgroup with a high-quality reference file is present, providing information on confident ancestral allelele calls.
The minor (folded) SFS uses the Minor Allele Frequency to calculate the Site Frequency and is folded against itself. This method is favourable if no closely related, high quality reference can be provided.
Input Data
There are two types of input data for both SFS types (derived and minor):
VCF: Suitable for medium to high coverage genomes (~ above 5X), straightforward generation of 2dSFS with easySFS.
Genotype Likelihoods: Can be inferred on low coverage data by NGS tools such as ANGSD. This workflow requires more steps, yet it can maximize information load from shallow genomes and retain more information.
Creating multidimensional SFS from VCF
To obtain multiple 2dSFS, we can use the tool easySFS. The name of this tool hints at the straight-forward process to generate the 2dSFS. It automates the pairing of multiple populations for all 2dSFS calculations, as needed for tools like fastsimcoal. In easySFS, you may simply provide a VCF file with all focal taxa and a tab-delimited population file with the two columns SAMPLE POPULATION without a header.
Then run this command in your Linux CLI:
./easySFS.py -i YourVCF.vcf -p POPFILE.txt -a --preview
From this, you choose the sample size of each population (projection) that maximizes the number of seggregating sites. So for the example files wcs_1200 in the easySFS folder, we would choose a projection of 50,133(0,1) or 50,134(0,1). This scales up with additional generation. Afterwards, we can directly run the conversion to 2dSFS with the command:
./easySFS.py -i YourVCF.vcf -p POPFILE.txt -a --proj=0,1 --prefix PREFIX
If you have a suitable ancestral reference to polarize the ancestral alleles of your vcf, you can add the flag –-unfold for the inference of the unfolded SFS.
EasySFS will then create an output folder with multidimensional SFS. It even pre-formats the files for direct usage with fastsimcoal and dadi. Careful, easySFS omits invariant sites which are not saved in the VCF. Thus, they need to either be added manually or ignored during downstream analyses
Creating multidimensional SFS from ANGSD Genotype Likelihoods
Infer the Site Allele Frequency (SAF) files with ANGSD by this command:
angsd -nThreads 4 -bam "$file"_BAM.list -out OUT_DIR -anc ANC.fasta -rf CTG_FILE.list -DoSaf 1 -GL 2
A reference sequence is required in this step and only derived SFS should be generated in angsd. If you plan to calculate the folded spectrum afterwards, you can perform the folding in realSFS.
realsfs POP1.saf.idx -fold 1 > POP1.sfs
From the individual SAF files, you can directly call two-dimensional SFS with realSFS.
realsfs POP1.saf.idx POP2.saf.idx > 2dPOP1_2.sfs
If higher dimensionality is required, I would recommend to call 3dSFS or beyond in winSFS.
./winsfs POP1.saf.idx POP2.saf.idx POP3.saf.idx > 3dPOP1_2_3.sfs
