Introducing Allele Frequency Inference from Shallow Genomes
Calculating per-site allele frequency is a crucial element in population genomics as it provides insights into ancestry and deviations from Hardy-Weinberg Equilibria. However, the inference of allele states on shallow genome is challenging; stochastic phenomena, such as allelic dropout or false homozygosity obscure the estimation of allele frequencies within datasets. Particularly at 1-2X coverage depth, correctly determining heterozygous states remains a game of chance. Learn how to navigate these statistic uncertainties and harness the full power of Genotype Likelihoods to reliably infer allele frequencies on shallow genomes
BIOINFORMATICS
Tim L. Heller
6/8/20254 min read
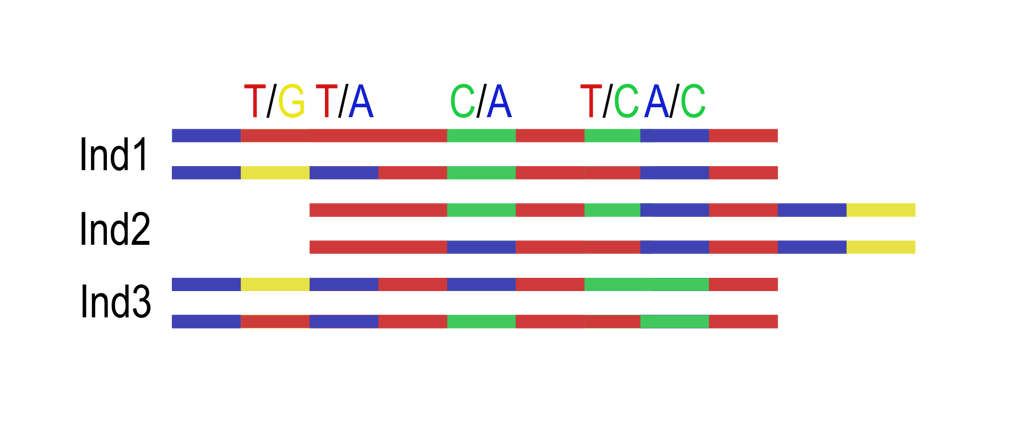
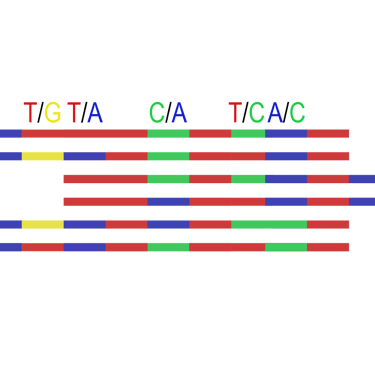
Calculating per-site allele frequency is a crucial element in population genomics as it provides insights into ancestry and deviations from Hardy-Weinberg Equilibria. However, the inference of allele states on shallow genome is challenging; stochastic phenomena, such as allelic dropout or false homozygosity obscure the estimation of allele frequencies within datasets. Particularly at 1-2X coverage depth, correctly determining heterozygous states remains a game of chance. Genotype Likelihoods offer an excellent solution to partially handle this uncertainty by assigning likelihoods to allele states rather than fixed calls. While this format is more computationally demanding, it can provide more accurate results. Nonetheless, despite its uprise there is only a limited selection of algorithms that can handle this data format. Developing an adequate understanding of how to infer and interpret per-site allele frequency with Genotype Likelihoods by tools like the ANGSD suite provides an excellent pathway to circumvent the dependency of hard-calls in software working plink or Variant call files (VCF) and unlocks the potential to program solutions fully independent from fixed call formats.
Harnessing the full potential of the ANGSD Suite
ANGSD (Analysis of Next Generation Sequencing Data) is a powerful toolset that can accompany your analysis of low-coverage genomes through the whole process. The initial stage is supplying mapping files (bam) and calling your Genotype Likelihoods (and other data formats). After this step, allele frequency estimation can be invoked by adding the -doMAF and -doMajorMinor flags and specifying their modes.
What are Major, Minor, and Ancestral Alleles?
These terms might appear self-explanatory, yet their use in ANGSD suite is flexible and define their role in allele state inference rather than observed biological phenomena. We could define these states as:
Major: The primary allele state, potentially the most frequent base at a site but definition can vary.
Minor: The alternative allele state of the major allele. The anker for the minor allele frequency metric.
Ancestral/Reference: The reference allele states, which can be included for comparative functions or serve certain roles. Only present when explicitly invoked with -ref or -anc; they are not automatically included otherwise.
Correctly Interpreting the -doMajorMinor flag.
This flag is important as it designates the roles of these allele types based on parameters. ANGSD uses different modes for this.
1: Genotype Likelihoods: Estimate Major and Minor alleles from Genotype Likelihoods. Major is the more common allele, Minor the alternative allele state.
2: Base Counts: Choosing the two most frequent bases across individuals by counts, NOT by Genotype Likelihoods.
3: Pre-defined: Designating Major and Minor allele by pre-defined files using the -sites option
4: Force reference major: Fixing the major allele (thus, not necessarily the most frequent allele) by the reference allele state of a fasta file using the -ref option. The minor (alternative) allele frequency is estimated by Genotype Likelihoods.
5: Force ancestral major: Fixing the major allele by the ancestral allele state of a fasta file using the -anc option. The minor (alternative) allele frequency is estimated by Genotype Likelihoods.
To my knowledge, there is no difference in allele frequency estimation between ancestral and reference alleles.
Applying the -doMAF option:
This option specifies, how you calculate the allele frequency themselves.
1: Using the knownEM algorithm to infer the minor allele frequency using the EM algorithms on Genotype Likelihoods .
2: Using the unknownEM algorithm to infer the minor allele frequency. Instead of specifying a single minor allele, ANGSD considers all three remaining alleles as potential minor alleles and sums over their probabilities (marginalizing) to obtain the minor allele frequency.
4: Calling the Minor Allele Frequency directly from the Genotype Probabilities supplying a beagle file (-beagle).
8: Directly inferring the allele frequency from simple base counts. Needs the -doCounts 1 option activated too.
These options can be further combined by adding the numbers. For example, knownEM and unknownEM are often combined (1 + 2) by -doMAF 3 to obtain both types of Minor Allele Frequencies
Inferring the Major Allele Frequency
As we are often assuming bi-allelic (ancestral vs. derived) states in many analyses and unknownEM further calculates the Minor Allele frequency by the summarized probability of non-major alleles, the major allele frequency can be approximated by
Major Allele Frequency ~ 1 – (unknown) Minor Allele Frequency.
Filtering Allele Frequencies
When calling allele frequencies, it is imperative to appropriately filter your sites and avoid artefacts. Relevant filters include:
-minMaf: Define a Minimum Allele Frequency from the -doMAF calculations. Helpful to include / exclude (near-)fixed sites.
-SNP_pval: Set a p-value for the allele state inference, hence insignificant estimation will get removed from the output.
-minInd: Remove sites with data in less than a given number of individuals.
-rmTriallelic: Remove tri-allelic sites from the output.
-setMinDepth: Set minimum coverage depth across all samples for more accurate allele state inference, requires -doCounts 1
-setMinDepthInd: Set minimum coverage depth per sample. Particularly useful in uneven coverage distribution, requires -doCounts 1
Interpreting the output files
The output files are gzipped in a .maf format. To read them, we can use the zcat function or gunzip them completely. The data structure is a tab separated file with the columns:
chromo: Contig on which the site was called.
Position: Base position of the site on the contig
Major: Major base allele, see above
Minor: Minor (alternative) base allele, see above.
knownEM: Allele frequency of the minor allele inferred by Genotype Likelihoods.
unknownEM: Allele frequency of the minor allele inferred by the probabilities of the non-major alleles.
pK-EM: Only present when -SNP_pval was defined. Calculated p-value of your knownEM allele frequency.
pu-EM: Only present when -SNP_pval was defined. Calculated p-value of your unknownEM allele frequency.
